Unix crash course
Familiarity with the Unix command-line is very likely the most foundational skillset we can develop for bioinformatics (and for many computational arenas beyond bioinformatics). This is a set of 5 introductory tutorials to help us get from being completely new to Unix up to being great friends with it 🙂
Some terminology
Here are some terms that are often used interchangeably – not because it’s important to remember them or any differences (it’s not for most of us), but just to have them laid out somewhere.
| Term | What it is |
|---|---|
command line |
a text-based environment capable of taking input and providing output |
shell |
what we use to talk to the computer; anything where we are pointing and clicking with a mouse is a Graphical User Interface (GUI) shell; something with text only is a Command Line Interface (CLI) shell |
Unix |
a family of operating systems |
bash |
the most common programming language used at a Unix command-line |
Why learn the Unix shell?
- enables the use of non-GUI (Graphical User Interface) tools
- reproducibility
- quickly perform operations on large files (without reading into memory)
- automation of repetitive tasks (need to rename 1,000 files?)
- enables use of higher-powered computers elsewhere (server/cloud)
- it’s the foundation for most of bioinformatics
Before we start
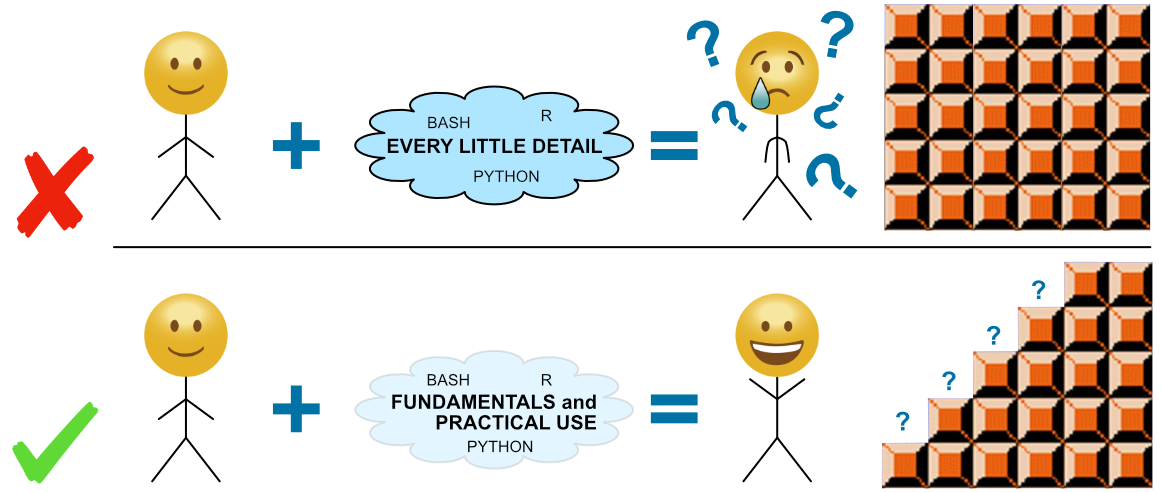
NOTE: Maybe the most imporant thing to keep in mind while going through these pages is that this is all about exposure, not memorization or mastering anything. Don’t worry about the details! Starting to build a foundation of fundamentals will allow us to figure out more details for specific things when we need to 🙂